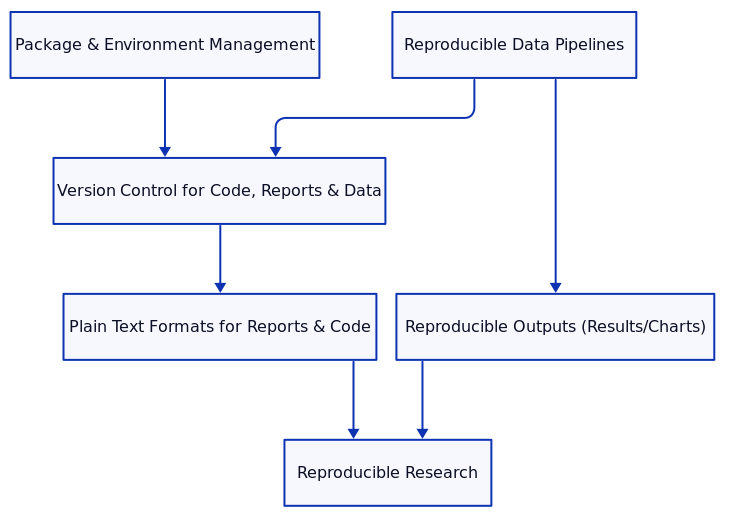
A Modular Approach to Reproducible Research
Reproducibility is a cornerstone of scientific progress, ensuring that research results can be replicated and verified by others, as well as by the original researchers at a later time. By breaking down research practices into distinct modules, researchers can address key challenges that arise in achieving reproducibility. This blog covers essential modules for ensuring reproducibility: package and environment management, reproducible data pipelines, version control for code, reports, and data, reproducible output, and the advantages of plain text formats.
1. Package Version and Environment Management
Managing package versions and environments ensures that your code runs identically across different systems, independent of updates or changes in dependencies. Tools such as Mamba, Pixi, and UV allow researchers to create isolated environments that lock down specific versions of software libraries and tools.
Why Environment Management?
Reproducibility breaks down when code that once worked suddenly fails due to updated dependencies. By using environment managers, you can “freeze” the environment configuration so others can recreate it exactly.
Example: Using Mamba for Environment Management
# Create an environment with specific package versions
mamba create -n research_env python=3.9 numpy=1.21 pandas=1.3 matplotlib=3.4
# Activate the environment
mamba activate research_env
# Export the environment configuration
mamba env export > environment.yml
# Recreate the environment from YAML
mamba env create -f environment.ymlBy sharing the environment.yml file, anyone can recreate the same environment, ensuring that code runs identically.
2. Reproducible Data Pipelines
After setting up the environment, the next step is ensuring that data processing pipelines produce consistent results. A reproducible data pipeline ensures that given the same raw data and the same processing steps, the outputs will be the same.
Why Reproducible Data Pipelines?
Data manipulation, cleaning, and transformation steps can introduce variability if not well-structured. Using pipeline automation tools such as Snakemake or Nextflow allows researchers to define and automate the data processing workflow.
Example: Data Pipeline with Snakemake
# Example rule in Snakemake for processing data
rule process_data:
input:
"data/raw_data.csv"
output:
"data/processed_data.csv"
shell:
"python process_data.py {input} {output}"By defining each step of the data processing in a reproducible pipeline, researchers can ensure consistent results across multiple runs.
Best Practices for Reproducible Pipelines:
- Explicit Inputs and Outputs: Always define clear inputs and outputs for every step.
- Automation: Use tools that automate dependencies and execution orders, reducing the chance for errors.
- Version Control Pipelines: Store pipeline scripts in a version control system to track changes over time.
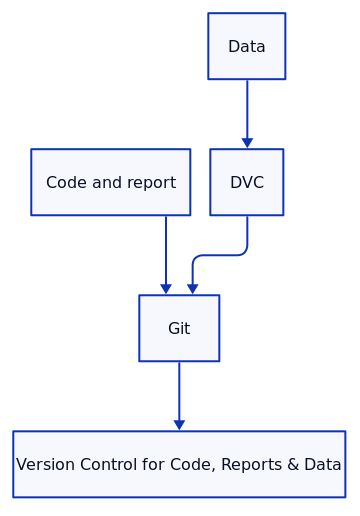
3. Version Control for Code, Reports, and Data
Version control is essential for tracking changes in code, data, and reports. Tools like Git for code and documents, and Data Version Control (DVC) for large datasets, help ensure that each aspect of your research can be traced and reproduced at any point in time.
Version Control for Code and Reports:
By using Git for version control, researchers can track changes in code and reports, ensuring that every version of the research is preserved. This also enables collaboration, as multiple contributors can work on the same project simultaneously.
Example: Version Control for Code with Git
# Initialize a Git repository
git init
# Add your code, environment, and report files
git add script.py environment.yml report.tex
# Commit your changes
git commit -m "Initial commit with code, environment, and report"Data Version Control with DVC:
When working with large datasets, DVC can help version your data and link it to specific versions of your code. This is crucial for machine learning projects where training data may evolve over time.
Example: Version Control for Data with DVC
# Initialize DVC in your project
dvc init
# Track a large dataset
dvc add data/raw_data.csv
# Add the data version to Git
git add data/raw_data.csv.dvc
git commit -m "Add versioned data"This setup ensures that both code and data can be versioned together, so changes in data are linked to specific code versions.
4. Reproducible Output: Results and Charts
With the environment and data pipeline set up, the next focus is ensuring that the research output — whether it’s results, figures, or charts — is consistent. Reproducibility in output guarantees that the same results are obtained when the analysis is rerun.
Why Reproducible Output?
Charts, tables, and numerical results should be consistent across different machines or different runs of the code. This is especially important when randomness is involved in the process (e.g., random sampling or bootstrapping).
Example: Generating Reproducible Charts in Python
import numpy as np
import matplotlib.pyplot as plt
# Set a seed for reproducibility
np.random.seed(42)
# Generate random data
data = np.random.randn(1000)
# Create a histogram
plt.hist(data, bins=30, color='blue', alpha=0.7)
plt.title("Reproducible Histogram")
plt.savefig("reproducible_histogram.png")Ensuring that random seeds are set, using standardized file formats, and specifying output directories are key practices for maintaining reproducibility.
5. Plain Text Formats for Version Control
When it comes to version control, using plain text formats like LaTeX/Typst for reports and .py for code is far superior to other formats like .docx (Microsoft Word) which uses XML for representing the document internally or Jupyter notebooks which uses JSON. Such formats can be difficult to manage in version control systems like Git.
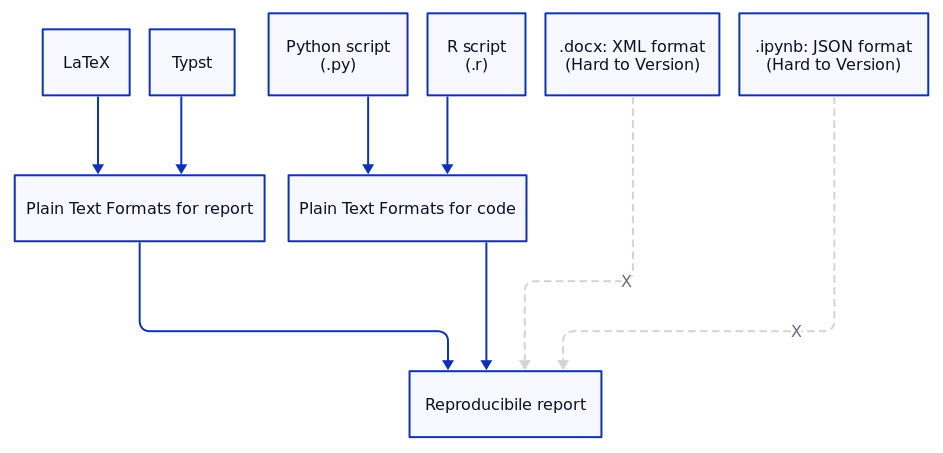
Why Plain Text (LaTeX/Typst) is Better Than .docx or Google Docs:
- Clear Diffs and Merges: Plain text formats allow Git to track changes at a line-by-line level. With formats like
.docx, changes are harder to track, and merging conflicts is more difficult. - Consistency: LaTeX and Typst ensure that the document formatting, equations, and citations remain identical across different machines and compilers.
- Lightweight: Plain text files are smaller and easier to manage, whereas
.docxfiles can be bulky and prone to corruption.
Why Plain Text .py Files are Better Than Jupyter Notebooks:
- Clear Execution Order: Jupyter notebooks hide the state of the execution, leading to inconsistent results if cells are run out of order. Plain
.pyfiles, in contrast, provide a linear flow, making them easier to debug and reproduce. - Simpler Version Control: Jupyter notebooks are stored as JSON, which makes diffing and merging difficult. Plain text
.pyscripts, however, are perfectly suited for Git-based version control.
Example: Converting Jupyter Notebook to .py
# Convert a Jupyter notebook to a plain text Python script
jupyter nbconvert --to script notebook.ipynbUsing .py files over Jupyter notebooks enhances reproducibility by making the code clearer, more traceable, and easier to integrate into a larger workflow.
Conclusion
By adopting a modular approach to reproducible research, you can ensure that every aspect of your work—environment, data pipeline, code, results, and reports—can be replicated by others. Starting with environment management, followed by reproducible data pipelines, and finally tracking everything with version control, researchers can create transparent and robust workflows.
Using plain text formats like .py, LaTeX, and Typst enhances version control and ensures that code, data, and documents remain lightweight and version-traceable. Integrating these practices into your research guarantees that your work can stand the test of time and be built upon by the wider scientific community.

Leave a Reply
Want to join the discussion?Feel free to contribute!